Add More Variables
add_more_variables.RmdIf you want to use add variables from the original medicare files to all medicare years, like MedPAR or Carrier, this article would help you to do so.
Why add more variables?
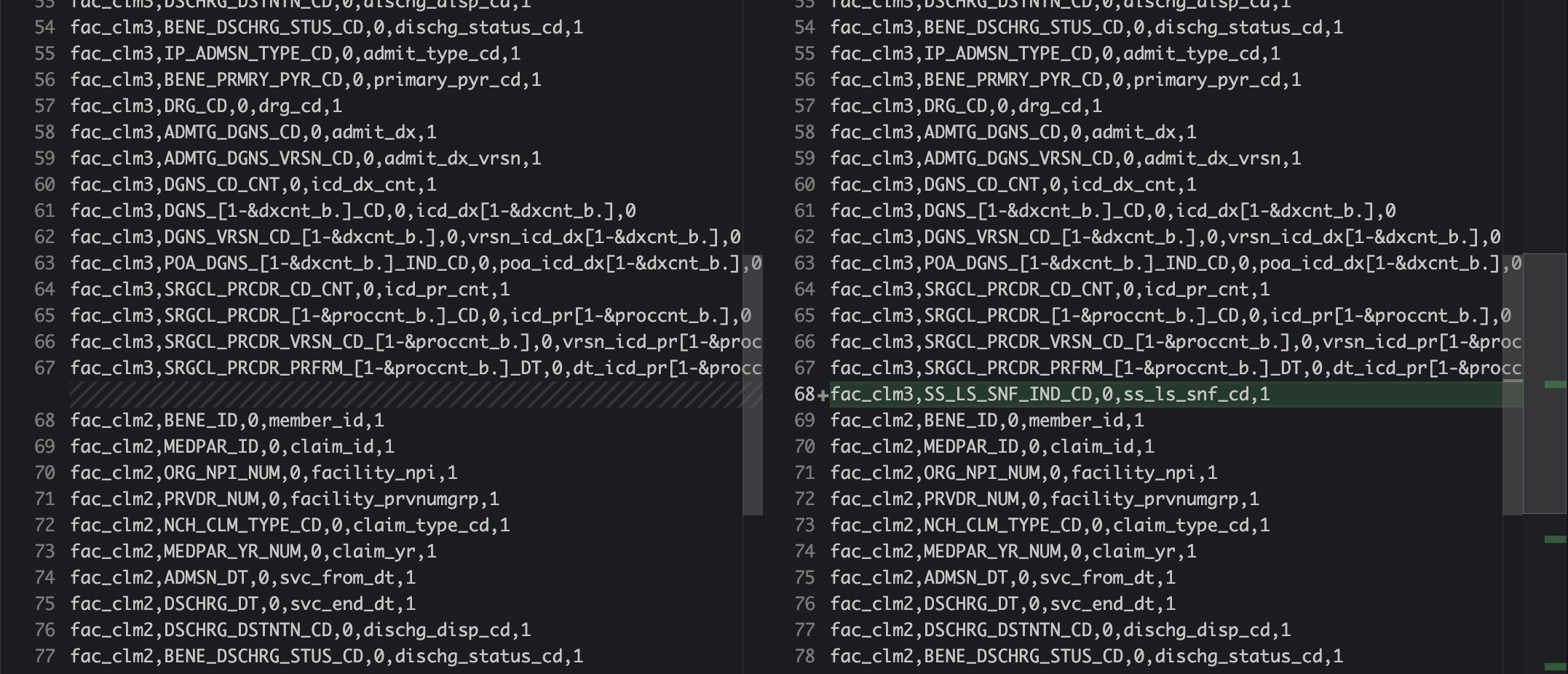
You have learned more about medicare and found some useful variables that were not previously used in medicare standardization steps. For example, I recently learned from ResDAC that MedPAR has a variable SS_LS_SNF_IND_CD to indicate if it’s SNF stay. This is particularly useful since our research is about inpatient procedures. So we would want to exclude SNF claims. I wanted to add this new var to all medicare standardized MedPAR for our analytic files.
How to add more variables
Step1: select vars from SAS files
Since all our original files are in SAS format, first step to add this new variable, for example SS_LS_SNF_IND_CD, to our variable selections from SAS
the sas code to do so can be found in
medicare_std/code/other/sas_code/create_0.1perc_sample_from_sas_data/Medpar.sas
# sas code example
proc export data = medpar.medpar2007 (keep = ORG_NPI_NUM BENE_ID MEDPAR_ID PRVDR_NUM ADMSN_DT DSCHRG_DT DSCHRG_DSTNTN_CD BENE_DSCHRG_STUS_CD IP_ADMSN_TYPE_CD BENE_PRMRY_PYR_CD DRG_CD ADMTG_DGNS_CD DGNS_CD_CNT SRGCL_PRCDR_CD_CNT bene_id DGNS_1_CD DGNS_2_CD DGNS_3_CD DGNS_4_CD DGNS_5_CD DGNS_6_CD DGNS_7_CD DGNS_8_CD DGNS_9_CD DGNS_10_CD SRGCL_PRCDR_1_CD SRGCL_PRCDR_2_CD SRGCL_PRCDR_3_CD SRGCL_PRCDR_4_CD SRGCL_PRCDR_5_CD SRGCL_PRCDR_6_CD SRGCL_PRCDR_PRFRM_1_DT SRGCL_PRCDR_PRFRM_2_DT SRGCL_PRCDR_PRFRM_3_DT SRGCL_PRCDR_PRFRM_4_DT SRGCL_PRCDR_PRFRM_5_DT SRGCL_PRCDR_PRFRM_6_DT SS_LS_SNF_IND_CD)
dbms=csv
outfile="Y:\original_medicare_selected_vars\data\MedPAR\medpar2007.csv"
replace;
run;Step2: create 0.1% data from the new csv files
code is at
"medicare_std/code/other/sample_raw_data/sample_raw_medicare.R"
# code from the file
# DO NOT RUN
# select medpar records related to the bene in first step
medpar_data_dir <- create_path(
medicare_dir = medicare_dir,
data_folder_name = medpar_folder
)
medpar_list <- list(
demon_sample_data_path = demon_data_dir$sample_data_path,
read_full_data_path = medpar_data_dir$full_data_path,
save_sample_data_path = medpar_data_dir$sample_data_path
)
no_cores <- availableCores() - 2
plan(multicore, workers = no_cores)
future_pmap(medpar_list, select_sample, .progress = TRUE)